Dev:Subform-block BiometryIOL700: Difference between revisions
Jump to navigation
Jump to search
No edit summary |
No edit summary |
||
| (37 intermediate revisions by 3 users not shown) | |||
| Line 14: | Line 14: | ||
!Comment | !Comment | ||
!Formula | !Formula | ||
! | !XML import | ||
|- | |||
|Link | |||
|Import | |||
|Importieren | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
| | |||
|- | |- | ||
|Dropdown | |Dropdown | ||
| Line 27: | Line 40: | ||
|Show all device instances of Device type - BIO | |Show all device instances of Device type - BIO | ||
| | | | ||
| | |</XmlVersion><DeviceVersion> | ||
<ProductName> | |||
|- | |- | ||
|Time box | |Time box | ||
| Line 40: | Line 54: | ||
| | | | ||
| | | | ||
| | |</Patient> | ||
<AcquisitionTime> | |||
|- | |- | ||
|Dropdown | |Dropdown | ||
| Line 52: | Line 67: | ||
| | | | ||
| | | | ||
| | |Phakic, Aphakic, Pseudophakic, Phakic IOL, Piggyback IOL, Piggyback IOL silicone, Pseudophakic PMMA, Pseudophakic silicone | ||
| | |</Patient> | ||
<LensState> | |||
|- | |- | ||
|Dropdown | |Dropdown | ||
| | |LVC | ||
| | |LSK | ||
| | |BIO | ||
| | |LVCST | ||
| | |BIO | ||
| - | | - | ||
| | | | ||
| Line 66: | Line 82: | ||
|Lens State (treated, untreated- refsurgery) | |Lens State (treated, untreated- refsurgery) | ||
TS: Laser Vision Correction/ Augenlaserkorektur [myopic,hyporopic] | TS: Laser Vision Correction/ Augenlaserkorektur [myopic,hyporopic] | ||
| | |Untreated, LASIK, LASEK, PRK, RK | ||
| | |<Patient> | ||
<RefractiveSurgery> | |||
|- | |- | ||
|Dropdown | |Dropdown | ||
| | |VBS | ||
| | |GS | ||
|VIT | |VIT | ||
|VBS | |VBS | ||
|BIO | |BIO | ||
| Line 82: | Line 96: | ||
| | | | ||
|Vitreous Body State | |Vitreous Body State | ||
| | |Vitreous body, Silicone oil, Post vitrectomy | ||
| | |<Patient><VitreousBodyState> | ||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 91: | Line 105: | ||
|SPH | |SPH | ||
|BIO | |BIO | ||
| 0.25 | |0.25 | ||
| | | | ||
| | | | ||
| Line 117: | Line 118: | ||
|AXL | |AXL | ||
|BIO | |BIO | ||
| 0.01 | |0.01 | ||
| | | | ||
| | | | ||
|How to display status? | |||
| | | | ||
|</Patient> | |||
<Al> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
|ACD [mm] | |ACD [mm] | ||
|EXT VKT [mm] | |EXT VKT [mm] | ||
|BIO | |BIO | ||
|ACD | |ACD | ||
|BIO | |BIO | ||
| 0.01 | |0.01 | ||
| | | | ||
| | | | ||
|How to display status? | |||
| | | | ||
|</Patient> | |||
<Acd> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| | | AQD [mm] | ||
|INT VKT [mm] | |INT VKT [mm] | ||
|BIO | |BIO | ||
|AQD | |AQD | ||
|BIO | |BIO | ||
| Line 146: | Line 149: | ||
| | | | ||
| | | | ||
| | |Make sure that units are4 converter, CCT should be in mm in calculation | ||
|ACD-CCT | |ACD-CCT | ||
| | | | ||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 154: | Line 157: | ||
|LD [mm] | |LD [mm] | ||
|BIO | |BIO | ||
|LNT | |LNT | ||
|BIO | |BIO | ||
|0.01 | |0.01 | ||
| Line 160: | Line 163: | ||
| | | | ||
|Lens thickness | |Lens thickness | ||
How to display status? | |||
| | | | ||
| | |</Patient> | ||
<Lt> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| | |CRW1 | ||
| | |CRW1 | ||
| | |BIO | ||
| | |CRW1 | ||
| | |BIO | ||
| | |0.01 | ||
| | | | ||
| | | | ||
| | | | ||
|100 * ((AL/K)'4 - (AL/TK)'4) + CCT score + K score + 0.22 | |||
(please see below scr for CCT and K score values) | |||
| | | | ||
|- | |- | ||
|Number textbox | |Number textbox | ||
|CCT [µm] | |CCT [µm] | ||
|ZHD [µm] | |ZHD [µm] | ||
|TOMO | |TOMO | ||
|CCT | |CCT | ||
|BIO | |BIO | ||
| Line 186: | Line 192: | ||
|<400 | |<400 | ||
|Central corneal thickness | |Central corneal thickness | ||
How to display status? | |||
| | | | ||
| | |</Patient> | ||
<Cct> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
|R1 [mm] | |R1 [mm] | ||
|R1 [mm] | |R1 [mm] | ||
|BIO --> KER | |BIO --> KER | ||
| Line 200: | Line 208: | ||
|Maximal Radius | |Maximal Radius | ||
| | | | ||
| | |<Keratometry><MaximalRadius> | ||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 206: | Line 214: | ||
|K1 [Dpt] | |K1 [Dpt] | ||
|BIO --> KER | |BIO --> KER | ||
|K1(dim1) | |K1(dim1) | ||
|BIO | |BIO | ||
|1 | |1 | ||
| | | | ||
|>48 | |>48 | ||
| | |Check if calculation matches XML | ||
|332/R1 or 337.5/R1 | Formula should be implemented, XML import data is for checking only | ||
| | |332/R1 (if KIndex is 1.332) | ||
or | |||
337.5/R1 (if KIndex is 1.3375) | |||
|<Keratometry> | |||
<FlatK> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 226: | Line 238: | ||
| | | | ||
| | | | ||
| | |<Keratometry> | ||
<MaximalAxis> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
|R [mm] | |R mean [mm] | ||
|R [mm] | |R mean [mm] | ||
|BIO --> KER | |BIO --> KER | ||
|KR(dim1) | |KR(dim1) | ||
| Line 239: | Line 252: | ||
|Radius | |Radius | ||
|((R1+R2)/2) | |((R1+R2)/2) | ||
| | | | ||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 252: | Line 265: | ||
|Minimal Radius | |Minimal Radius | ||
| | | | ||
| | |<Keratometry> | ||
<MinimalRadius> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 259: | Line 273: | ||
|BIO --> KER | |BIO --> KER | ||
|K2(dim1) | |K2(dim1) | ||
|BIO | |BIO | ||
|1 | |1 | ||
| | | | ||
| | | | ||
| | | Check if calculation matches XML | ||
|332/R2 or 337.5/R2 | Formula should be implemented, XML import data is for checking only | ||
| | |332/R2 (if KIndex is 1.332) | ||
or | |||
337.5/R2 (if KIndex is 1.3375) | |||
|<Keratometry> | |||
<SteepK> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
|Axis [°] | |Axis [°] | ||
|Achse [°] | | Achse [°] | ||
|BIO --> KER | | BIO --> KER | ||
|K2(dim2) | |K2(dim2) | ||
|BIO | | BIO | ||
|1 | |1 | ||
| | | | ||
| Line 278: | Line 296: | ||
| | | | ||
| | | | ||
| | |<Keratometry> | ||
<MinimalAxis> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
|K [D] | |K mean [D] | ||
|K [Dpt] | |K mean [Dpt] | ||
|BIO --> KER | |BIO --> KER | ||
|KR(dim2) | |KR(dim2) | ||
| Line 290: | Line 309: | ||
| | | | ||
| | | | ||
| | |((K1 + K2)/2) | ||
| | | | ||
|- | |- | ||
|Dropdown | |Dropdown | ||
| Line 304: | Line 323: | ||
|Refractive index | |Refractive index | ||
| | | | ||
| | |<Keratometry> | ||
<RefractiveIndex> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 315: | Line 335: | ||
| | | | ||
| | | | ||
| | |Check if calculation matches XML | ||
|K1-K2 | Formula should be implemented, XML import data is for checking only | ||
| | |K1-K2 @ A1 | ||
|<Keratometry> | |||
<DeltaD> | |||
|- | |- | ||
|Number textbox | |Number textbox | ||
| Line 324: | Line 347: | ||
|BIO --> KER | |BIO --> KER | ||
|CAST(dim2) | |CAST(dim2) | ||
|BIO | |||
|1 | |||
| | |||
| | |||
| | |||
|Axis from K1 | |||
|<Keratometry> | |||
<DeltaAxis> | |||
|- | |||
|Button | |||
|Upload document | |||
|Dokument hochladen | |||
|BIO | |||
| | |||
|BIO | |||
| | |||
| | |||
| | |||
|Supports any file | |||
| | |||
| | |||
|- | |||
|Comment | |||
|Comment | |||
| Bemerkungen | |||
|BIO | |BIO | ||
| | |COMM | ||
| BIO | |||
| - | |||
| | |||
| | | | ||
| | | | ||
| | | | ||
| | | | ||
|} | |} | ||
==Mockup == | |||
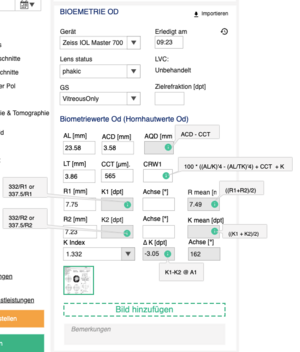
== Mockup == | [[File:Biometry block with formulas.png|left|thumb]] | ||
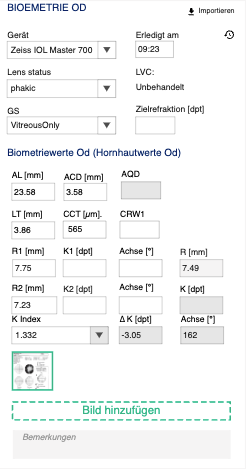
[[File:Biometry | [[File:Biometry block.png|left|thumb]] | ||
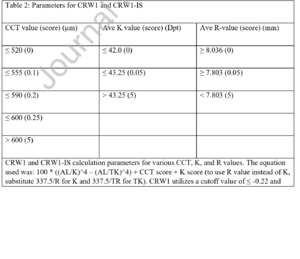
[[File:CRW1 formula.png|left|thumb]] | |||
Latest revision as of 13:57, 19 June 2023
Block fields
| Field type | Field name EN | Field name DE | Patient data set | Patient data item | Procedure | Step | Warning | Critical | Comment | Formula | XML import |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Link | Import | Importieren | |||||||||
| Dropdown | Device | Gerät | BIO | - | BIO | - | Show all device instances of Device type - BIO | </XmlVersion><DeviceVersion>
<ProductName> | |||
| Time box | Done at | Erledigt am | BIO | MESD | BIO | - | </Patient>
<AcquisitionTime> | ||||
| Dropdown | Lens status | Linsenstatus | BIO | LST | BIO | - | Phakic, Aphakic, Pseudophakic, Phakic IOL, Piggyback IOL, Piggyback IOL silicone, Pseudophakic PMMA, Pseudophakic silicone | </Patient>
<LensState> | |||
| Dropdown | LVC | LSK | BIO | LVCST | BIO | - | Lens State (treated, untreated- refsurgery)
TS: Laser Vision Correction/ Augenlaserkorektur [myopic,hyporopic] |
Untreated, LASIK, LASEK, PRK, RK | <Patient>
<RefractiveSurgery> | ||
| Dropdown | VBS | GS | VIT | VBS | BIO | - | Vitreous Body State | Vitreous body, Silicone oil, Post vitrectomy | <Patient><VitreousBodyState> | ||
| Number textbox | Target refraction [D] | Zielrefraktion [Dpt] | TREF | SPH | BIO | 0.25 | |||||
| Number textbox | AL [mm] | AL [mm] | BIO | AXL | BIO | 0.01 | How to display status? | </Patient>
<Al> | |||
| Number textbox | ACD [mm] | EXT VKT [mm] | BIO | ACD | BIO | 0.01 | How to display status? | </Patient>
<Acd> | |||
| Number textbox | AQD [mm] | INT VKT [mm] | BIO | AQD | BIO | 0.01 | Make sure that units are4 converter, CCT should be in mm in calculation | ACD-CCT | |||
| Number textbox | LT [mm] | LD [mm] | BIO | LNT | BIO | 0.01 | Lens thickness
How to display status? |
</Patient>
<Lt> | |||
| Number textbox | CRW1 | CRW1 | BIO | CRW1 | BIO | 0.01 | 100 * ((AL/K)'4 - (AL/TK)'4) + CCT score + K score + 0.22
(please see below scr for CCT and K score values) |
||||
| Number textbox | CCT [µm] | ZHD [µm] | TOMO | CCT | BIO | 1 | >=400 and <480 | <400 | Central corneal thickness
How to display status? |
</Patient>
<Cct> | |
| Number textbox | R1 [mm] | R1 [mm] | BIO --> KER | K1(dim3) | BIO | 1 | Maximal Radius | <Keratometry><MaximalRadius> | |||
| Number textbox | K1 [D] | K1 [Dpt] | BIO --> KER | K1(dim1) | BIO | 1 | >48 | Check if calculation matches XML
Formula should be implemented, XML import data is for checking only |
332/R1 (if KIndex is 1.332)
or 337.5/R1 (if KIndex is 1.3375) |
<Keratometry>
<FlatK> | |
| Number textbox | Axis [°] | Achse [°] | BIO --> KER | K1(dim2) | BIO | 1 | <Keratometry>
<MaximalAxis> | ||||
| Number textbox | R mean [mm] | R mean [mm] | BIO --> KER | KR(dim1) | BIO | 1 | Radius | ((R1+R2)/2) | |||
| Number textbox | R2 [mm] | R2 [mm] | BIO --> KER | K2(dim3) | BIO | 1 | Minimal Radius | <Keratometry>
<MinimalRadius> | |||
| Number textbox | K2 [D] | K2 [Dpt] | BIO --> KER | K2(dim1) | BIO | 1 | Check if calculation matches XML
Formula should be implemented, XML import data is for checking only |
332/R2 (if KIndex is 1.332)
or 337.5/R2 (if KIndex is 1.3375) |
<Keratometry>
<SteepK> | ||
| Number textbox | Axis [°] | Achse [°] | BIO --> KER | K2(dim2) | BIO | 1 | <Keratometry>
<MinimalAxis> | ||||
| Number textbox | K mean [D] | K mean [Dpt] | BIO --> KER | KR(dim2) | BIO | 1 | ((K1 + K2)/2) | ||||
| Dropdown | K Index | K index | BIO --> KER | REFIND | BIO | - | Refractive index | <Keratometry>
<RefractiveIndex> | |||
| Number textbox | ∆ K [D] | ∆ K [Dpt] | BIO --> KER | CAST(dim1) | BIO | 1 | Check if calculation matches XML
Formula should be implemented, XML import data is for checking only |
K1-K2 @ A1 | <Keratometry>
<DeltaD> | ||
| Number textbox | Axis [°] | Achse [°] | BIO --> KER | CAST(dim2) | BIO | 1 | Axis from K1 | <Keratometry>
<DeltaAxis> | |||
| Button | Upload document | Dokument hochladen | BIO | BIO | Supports any file | ||||||
| Comment | Comment | Bemerkungen | BIO | COMM | BIO | - |
Mockup